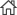-
- Prof. Kim Inki's Lab published in Nature communications
- Prof. Kim Inki's lab published 'Metasurface-driven full-space structured light for three-dimensional imaging' in Nature communications on October, 2022. Further details can be obtained from the following link. Kim, Gyeongtae, et al. "Metasurface-driven full-space structured light for three-dimensional imaging." Nature communications 13.1 (2022): 1-10.
-
- 작성일 2022-11-21
- 조회수 580
-

- Prof. Jang-Yeon Park's Lab published in Science
- Prof. Jang-Yeon Park published 'In vivo direct imaging of neuronal activity at high temporospatial resolution' in Science on October, 2022. Further details can be obtained from the following link. Toi, Phan Tan, et al. "In vivo direct imaging of neuronal activity at high temporospatial resolution." Science 378.6616 (2022): 160-168.
-
- 작성일 2022-10-18
- 조회수 687
-
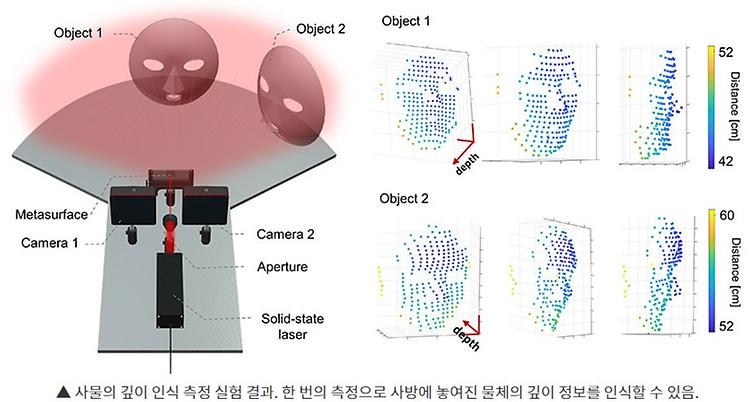
- Prof. Park Chun Gwon published in Biomaterials
- Prof. Park Chun Gwon published 'Image-guided in situ cancer vaccination with combination of multi-functional nano-adjuvant and an irreversible electroporation technique' in Biomaterials on October, 2022. Further details can be obtained from the following link. Press article : http://www.lecturernews.com/news/articleView.html?idxno=106458 Han, Jun-Hyeok, et al. "Image-guided in situ cancer vaccination with combination of multi-functional nano-adjuvant and an irreversible electroporation technique." Biomaterials 289 (2022): 121762.
-
- 작성일 2022-10-13
- 조회수 553
-
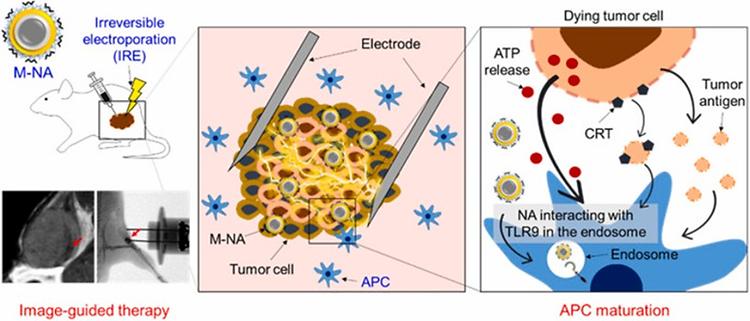
- Prof. Woo Choong-Wan's Lab published in Science Advances
- Prof. Woo Choong-Wan's Lab published 'When self comes to a wandering mind : Brain representations and dynamics of self-generated concepts in spontaneous thought' in Science Advances on August 31th, 2022. Further details can be obtained from the following link. Kim, Byeol, et al. "When self comes to a wandering mind: Brain representations and dynamics of self-generated concepts in spontaneous thought." Science advances 8.35 (2022): eabn8616.
-
- 작성일 2022-10-13
- 조회수 752
-
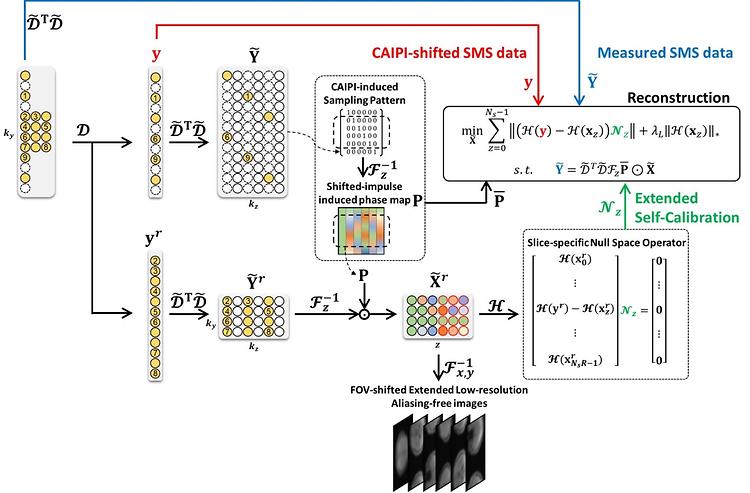
- Prof. Park Jaeseok's Lab published in Medical Image Analysis
- Prof. Park Jaeseok's Lab published 'Generalized self-calibrating simultaneous multi-slice MR image reconstruction from 3D Fourier encoding perspective' in Medical Image Analysis on November , 2022. Further details can be obtained from the following link. Lim, Eun Ji, et al. "Generalized self-calibrating simultaneous multi-slice MR image reconstruction from 3D Fourier encoding perspective." Medical Image Analysis (2022): 102621.
-
- 작성일 2022-10-13
- 조회수 782
-
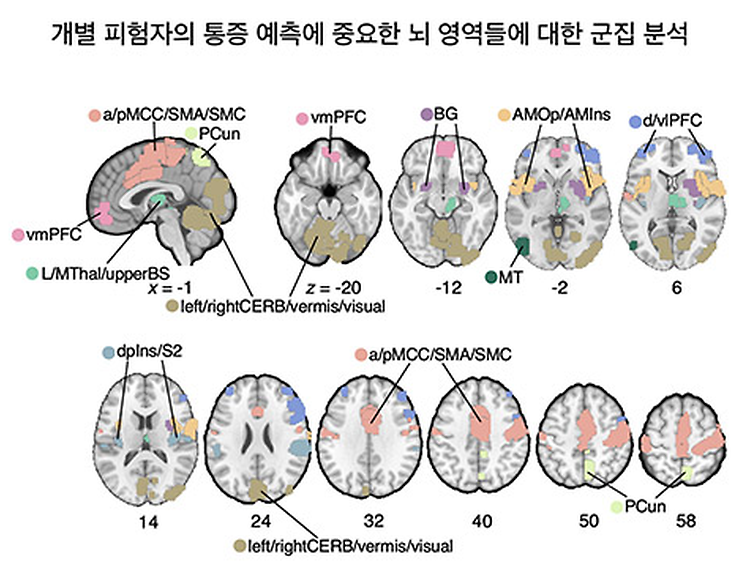
- Prof. Woo Choong-Wan's Lab published in Nature Neuroscience
- Prof. Woo Choong-Wan's Lab published 'Individual variability in brain representations of pain' in Nature Neuroscience on May 30th, 2022. Further details can be obtained from the following link. Kohoutová, L., Atlas, L. Y., Büchel, C., Buhle, J. T., Geuter, S., Jepma, M., ... & Woo, C. W. (2022). Individual variability in brain representations of pain. Nature Neuroscience, 1-11.
-
- 작성일 2022-06-15
- 조회수 692
-
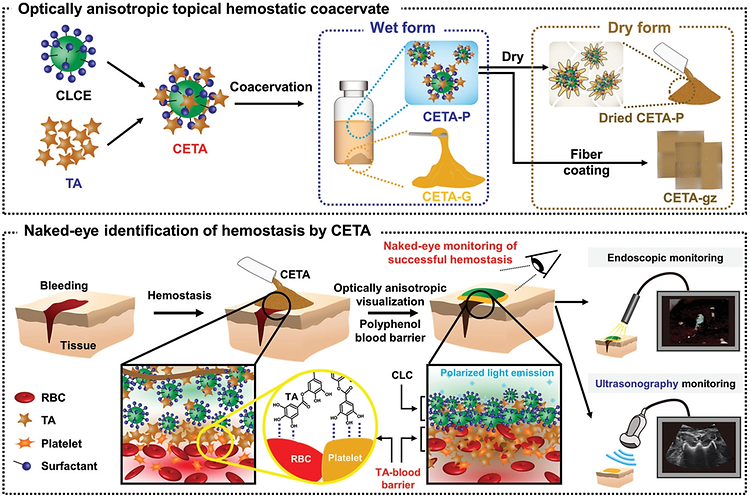
- Prof. Shin Mikyung's Lab publishes in Advanced Functional Materials
- Prof. Shin Mikyung's Lab published 'Optically Anisotropic Topical Hemostatic Coacervate for Naked-Eye Identification of Blood Coagulation' in Advanced Functional Materials on December 19th, 2021. Further details can be obtained from the following link. Jin, S., Kim, S., Kim, D. S., Son, D., & Shin, M. Optically Anisotropic Topical Hemostatic Coacervate for Naked‐Eye Identification of Blood Coagulation. Advanced Functional Materials, 2110320.
-
- 작성일 2021-12-24
- 조회수 593
-

- Prof. Chun Gwon Park wins the “Young Biomedical Engineering Award”
- Professor Chun Gwon Park won the Lutronic ‘Young Biomedical Engineering Award” at the Joint Conference of the IBEC and the ICBHI held on November 10th~12th. The ‘Young Biomedical Scholar Award’ is given to one of the scholars who has excellent industry-academic cooperation and academic achievements in the field of biomedical engineering, received his/her Ph.D. within 10 years and is below the age of 40. This award is sponsored by Lutronic, Korea’s No. 1 & World’s No. 10 laser medical device company. Professor Chun Gwon Park was selected as the winner for his contribution to the development of academic achievements and societies in the field of biomedical engineering and biomaterials. The Korean Society of Medical & Biological Engineering has more than 6,000 members and is a leading biomedical engineering/medical technology society in Korea since its foundation in 1979. Congratulations!
-
- 작성일 2021-11-23
- 조회수 644
-

- Lee Jae-Joong wins the KHBM Outstanding Trainee Award
- Lee Jae-Joong (Ph.D student) from Prof. Woo Choong-Wan's lab received "the KHBM Outstanding Trainee Award" at the 2021 fall workshop, organized by the Korean Society for Human Brain Mapping. The Korean Society for Human Brain Mapping(KHBM) was established in 2002 for the purpose of exchanging academic and technical information and building up Koreans characteristic brain mapping data on their own. It is a well-known academic association on brain function in Korea that leads research to study cognitive functions using cutting-edge imaging tools such as EEG/MEG, SPECT, PET and fMRI. The KHBM Outstanding Trainee Award is given to two people or under every year by evaluating the research achievements of the first author in the last year. Congratulations!
-
- 작성일 2021-11-12
- 조회수 439
-

- Prof. Woo Choong-Wan wins the KHBM Outstanding Achievement Award
- Prof. Woo Choong-Wan received the “the KHBM Outstanding Achievement Award” at the 2021 fall workshop, organized by the Korean Society for Human Brain Mapping. The Korean Society for Human Brain Mapping(KHBM) was established in 2002 for the purpose of exchanging academic and technical information and building up Koreans characteristic brain mapping data on their own. It is a well-known academic association on brain function in Korea that leads research to study cognitive functions using cutting-edge imaging tools such as EEG/MEG, SPECT, PET and fMRI. The KHBM Outstanding Achievement Award, the most prestigious award in the field of brain function mapping, is given to one person every year by comprehensively evaluating the research achievements of the lead author for last three years as a regular member of KHBM. Congratulations!
-
- 작성일 2021-11-12
- 조회수 461